Introductions:Cell-free DNA (cfDNA) are extracellular nucleic acids circulating in blood and other biological fluids. They are non-randomly fragmented, mainly short (< 200 bp) DNA molecules released as a result of apoptosis, necrosis or active secretion from cells. They are believed to be protected from degradation by nucleases due to nucleosome structure. cfDNA is a promising biomarker for prenatal testing, hemoglobinopathies, inflammatory progress, and detection of tumor-derived DNA in plasma. The amount of donor derived cell-free DNA circulating in the recipient's plasma can be used as a signature of dying cells from the transplanted organ. Donor derived cell-free DNA percent (%dd-cfDNA) can be quantified using next generation sequencing (NGS)-based method that utilizes genotype information of recipient and donor by measuring single nucleotide polymorphism (SNP) differences. This method was used in Genome Transplant Dynamics (GTD) cohort study. This study showed that elevated dd-cfDNA levels correlate with rejection of the transplanted organ. GTD method was shown to be a rigorous and highly reproducible across different genotyping or sequencing platforms. Here, we present thecfCloud, an open source, cloud-based implement of the GTD method in a Snakemake pipeline which automates the quantification of dd-cfDNA amount.
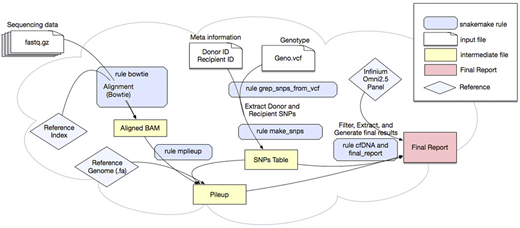
Methods:As illustrated in Figure 1, the analysis workflow contains several modules and rules; 1) Align reads rule - aligns sequencing reads against the reference genome (hg19), and marks duplicated reads. 2) Prepare genotype rule - given the input VCF file, for every recipient/donor pair, generates a SNP table by classifying SNPs as i) Group A, SNPs where both recipient (R) and donor (D) are homozygous with the same allele, like R=AA, D=AA, and ii) Group B, SNPs where both recipient and donor are homozygous with different alleles, like R=AA, D=BB. 3) Make CallsTable rule - for every SNP position, if the observed base of an overlapping read matches the recipient allele, it classifies that read as recipient-derived, if the observed base matches the donor allele then it classifies as donor-derived, otherwise as background-error. CallsTable lists chromosome, position, base quality and mapping quality scores of all the sites as well as the assigned category. 4) Call % dd-cfDNA rule - using the CallsTable, filters out the reads based on the input parameters. Then, calculates % dd-cfDNA by using SNPs in Group A and estimates background-error rate by using SNPs in Group B. 5) Prepare FinalReport rule - generates a final report showing total counts, donor counts, dd-cfDNA percentages and estimated background-error rates for all the samples.
Results:Using the workflow management system, Snakemake, we have developed a user friendly, efficient, and comprehensive pipeline that is fully automated, reproducible, and scalable. While the pipeline is fully customizable, it can be run using a simple command line. Upon execution, Snakemake infers the combination of rules necessary to achieve a specific output and or a report. With a basic understanding of the underlying framework of Snakemake, cfCloud is highly scalable and can be easily customized. In addition to local implementation, cfCloud also runs as an Amazon Machine Instance (AMI) in the Amazon Elastic Compute Cloud (EC2). The operating system, software tools and all settings, are encapsulated into a single cloud image of the computing system that is ready to deploy and can be easily saved and restored for later use. A snapshot is also an executable machine image that can be shared with other users of the cloud, allowing collaborators to share their configurations and analysis results as a single image.
Conclusions:Here we present a fully automated pipeline to systematize the quantification of donor derived cell-free DNA amount. The Amazon Web Machine Image is available from the AWS Market Place. The cfCloud provides users with improved usability, efficiency, and scalability in local and cloud environment. The cfCloud software, installation instructions, tutorials and example data can be freely obtained from https://github.com/NHLBI-BCB/cfCloud under the MIT license.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal